Genetic Linkage in Yeast
Identification and measurement of genetic linkage in yeast using tetrad dissection.
What is genetic linkage?
Genetic linkage is a technique for studying gene associations. The basic idea is that genes located close together on a chromosome are more likely to be inherited together. Using tetrad dissection, researchers can systematically study the products of meiotic cell division in yeast to reveal inheritance patterns that help identify regions of the genome where genes associated with a particular trait are likely to be located.
Tetrad dissection has been used to study genetic linkage for many decades. It remains a mainstay in modern yeast labs and a starting point for many classical genetic experiments, making it an important application to master for any budding yeast researcher.
Understanding genetic linkage:
Two genes are said to be ‘genetically linked’ if their phenotypic traits tend to be inherited together through generations. Microbiologists can deduce genetic linkage by studying the frequency of recombination during meiotic cell division in model organisms such as yeast.
Recombination is a process by which genetic material from either parent is reshuffled, resulting in new combinations of alleles in the offspring that are not present in either parent. It plays a big part in natural variation, generating novel genetic diversity within a population through the exchange of genetic material d between non-sister parent chromatids (two pairs of chromosomes) during cell division.
This genetic exchange occurs during ‘crossing over’, when pairs of chromosomes that share sequence homology become temporarily fused and swap segments of DNA. In yeast, the ability to easily identify the four spores from the same meiotic event allows the easy analysis of the products of recombination from crossing over.
A pair of genes that are located closely on the same chromosome are less likely to be separated by recombination than genes located at opposite ends of a chromosome. Thus by observing the segregation patterns of different phenotypes (traits) across the tetrad, researchers can infer the genetic linkage between the genes responsible for those traits.
How to identify genetic linkage:
Once the parental strains are mated and the tetrads dissected, the scientist observes the phenotype of each isolated spore. For each set of four spores, the phenotypes indicate a parental, non-parental or tetratype recombination event, as indicated in Figure 1.
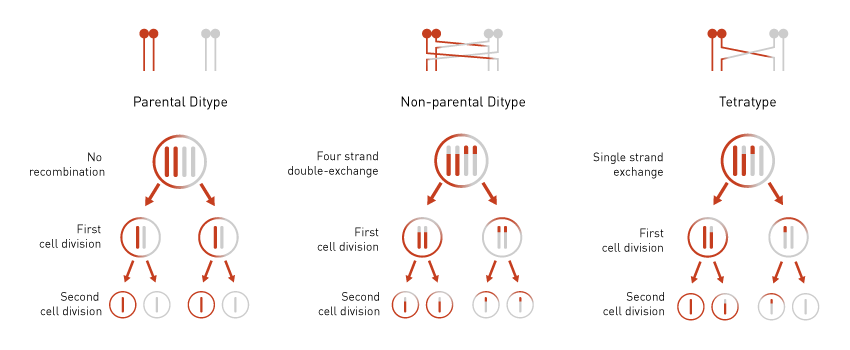
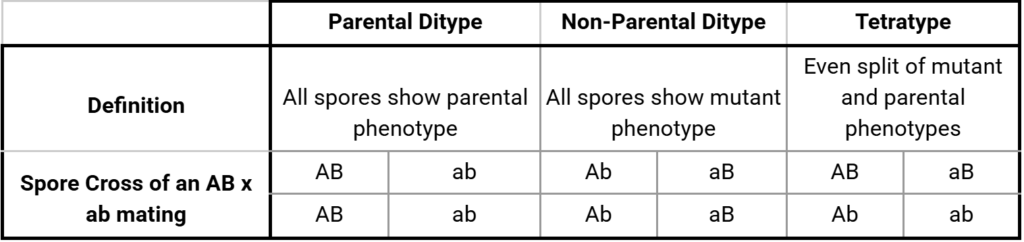
Figure 1: An example of tetrad analysis
The ratio of recombination types resulting from the same mating cross indicates the degree of genetic linkage between the genes of interest (Table 1). These ratios also indicate if one of the genes is located near the centromere where recombination is less likely to occur.
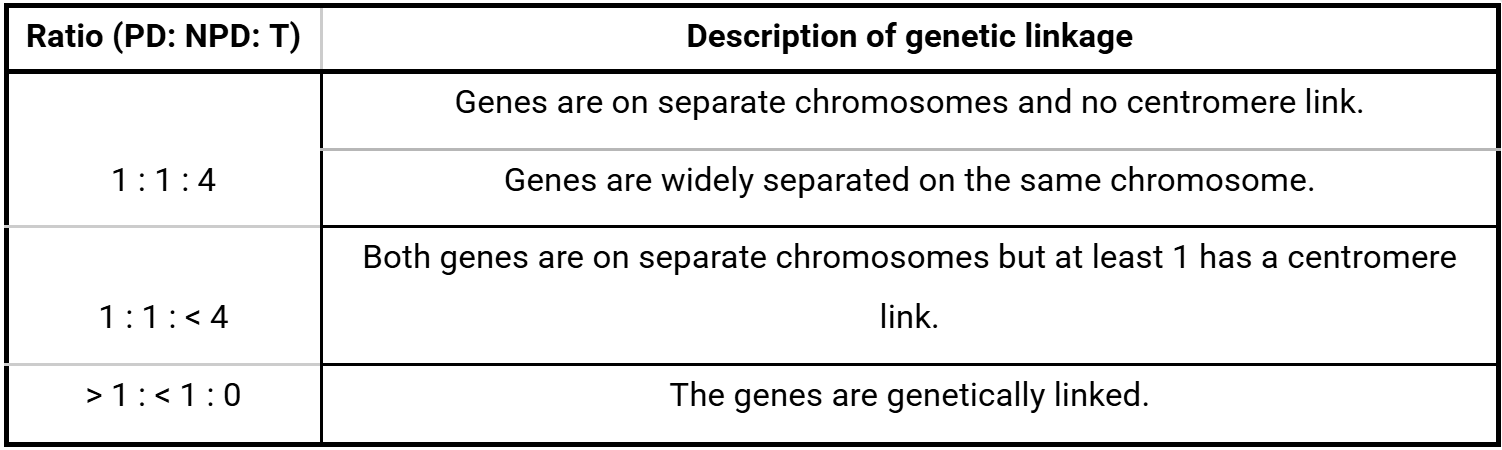
Table 1: How the ratio of the parental ditype (PD), non-parental ditype (NPD) and tetratype (T) phenotype patterns describe genetic linkage between two genes.
Measuring the distance between linked genes:
Once a genetic link between two genes has been identified, the next step is to measure the strength of that link. Sherman, 2002, shows how to calculate the distance between the genes, in centimorgans (cM), through the equation in Figure 2A. However, this equation is only accurate up to 35 cM. If the resulting number is greater than 35 cM,a correction is required following the equation, see Figure 2B. This data can also be used to calculate the distance between the gene and the centromere if a link was identified (refer to Table 1) using the equation shown in Figure 2C.
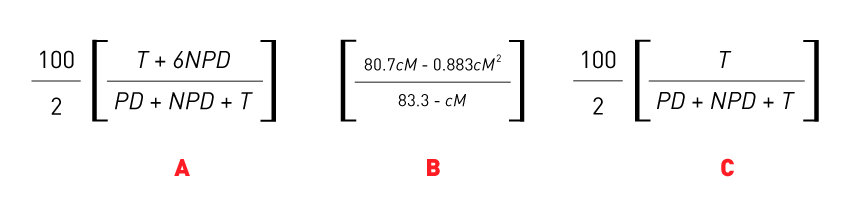
Figure 2: The series of equations used to (A) calculate the distance in centimorgans (cM) between two genes, (B) correct the equation in A if the distance is greater than 35 cM and (C ) calculate the distance from a centromere linked gene and the centromere, also known as cM’. T, PD and NPD can be replaced with the number of tetrads that displayed the tetratype, parental ditype or non-parental ditype pattern respectively, as described in Figure 1.
Want to dissect tetrads?
Discover the perfect fit for your needs from our selection of precision robots

Fiona Kemm MRes | Scientist
Fiona is a vital member of our Research team, rigorously testing our robots to ensure scientists don’t break them. With no prior robotics experience, she was the ideal guinea pig for our world-class user experience and support. Holding a BSc in Biochemistry and an MRes in Molecular Microbiology, Fiona brings extensive hands-on expertise she applies across departments, supporting both users and internal teams. From writing insightful web articles to specialising in SQWERTY, Fiona ensures our innovations perform flawlessly, helping customers focus on the creative and interpretive aspects of science that can’t be automated.